12.24.2025
Announcing ADAMS
Agent-Driven Autonomous Molecular Simulations
By The Rhizome Team
Introduction
At Rhizome, we recognize both the fundamental limitations and strengths of LLMs, and we focus our energy on where they excel. LLMs are polymathic knowledge bases with stellar coding capabilities, making them fundamental to the next generation of innovation in computational chemistry and biology, including small molecule discovery. That is why today we are announcing the open source release of ADAMS: Agent-Driven Autonomous Molecular Simulations.
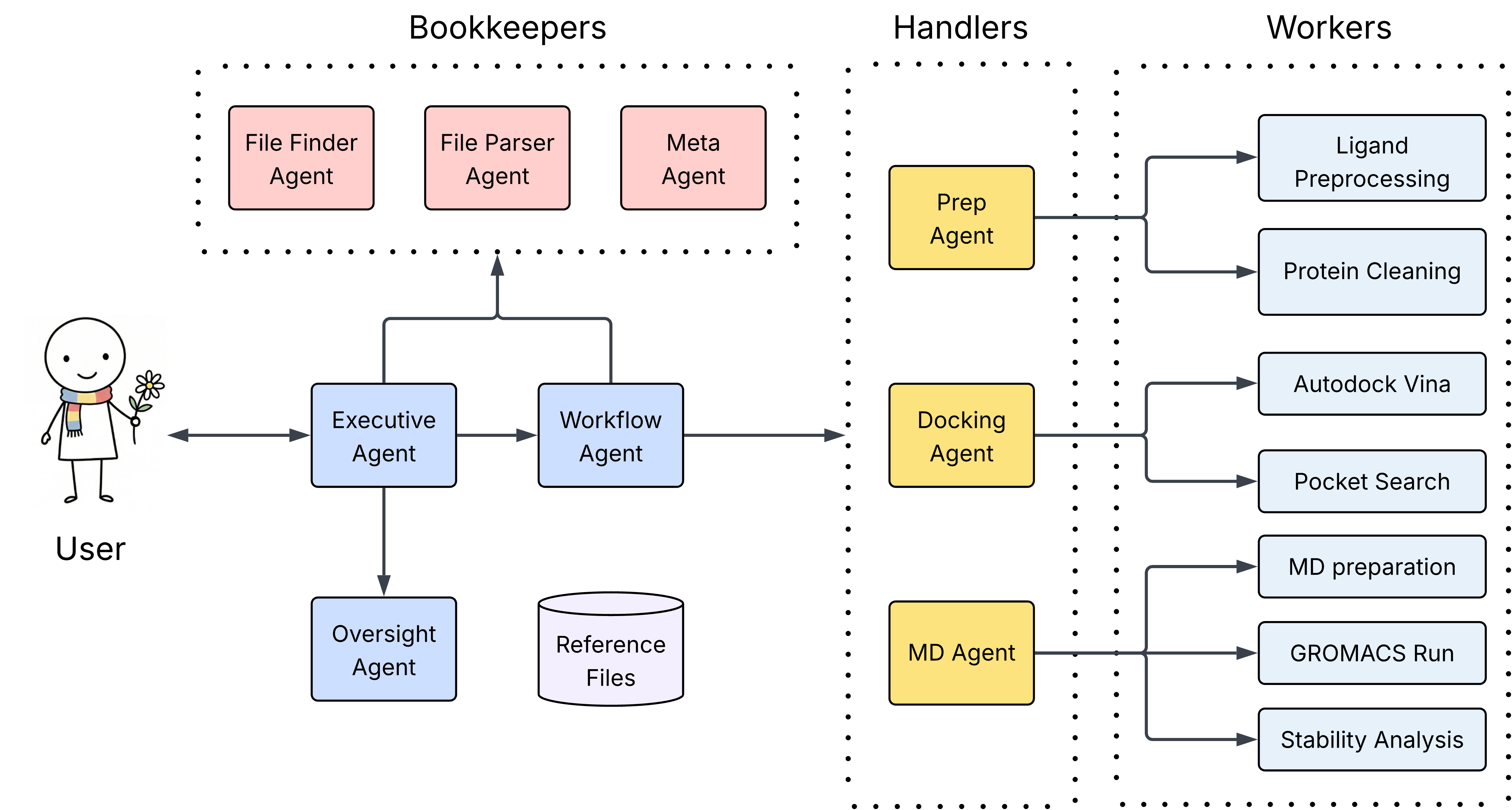
Letting you describe your docking and stability analysis in natural language - ADAMS handles protein preparation, binding site discovery, molecular docking, and MD simulations automatically. What used to require expert knowledge of multiple tools and days of scripting now takes a single command. Run end-to-end with a single command or drive individual stages for advanced control.
ADAMS
Installation is easy:
# One-command install
curl -fsSL https://raw.githubusercontent.com/Applied-Scientific/ADAMS/main/scripts/install.sh | bash
# Or download and inspect first
curl -fsSL https://raw.githubusercontent.com/Applied-Scientific/ADAMS/main/scripts/install.sh -o install.sh
bash install.shThe installer creates a conda environment with Python 3.12 and all dependencies (rdkit, openbabel, openmm, AmberTools, GROMACS). Set your OpenAI API key and you're ready to go.
How to Use
ADAMS uses a conversational interface. Launch it from your data directory:
adamsThen describe what you want to accomplish:
> Dock the ligands from Custom_Library.xlsx to 2PPN protein and run MD simulations on the top 3 binding poses
The system can: clean the protein structure (remove water, add hydrogens), process the ligand library (validate SMILES, filter by molecular weight), discover binding pockets across the protein surface, perform production docking at the top sites, run molecular dynamics simulations on the best poses and generate stability analysis reports. All the output is organized in timestamped directories such as agent_data/outputs/run_20251224_143022.
ADAMS scans the working directory and any additional folders specified by the user to discover available inputs and determines the appropriate starting point. Simply by talking to ADAMS, a fully customized plan can be created to start from any stage in the pipeline. The agents are capable of following the user prompt to perform customizable data pre-processing, docking and MD simulations. The user no longer needs to follow a rigid workflow or worry about strict file formatting. This makes it easy to integrate ADAMS into existing workflows or resume from partial results.
While long MD simulations can take hours or days, ADAMS tracks execution state in trace files and can resume from any stage:
adams
> Resume the run from yesterday that was interrupted during MD analysis
The meta-analysis agent reads trace files (agent_data/traces/trace_20251223_103022.jsonl) to determine where to continue. All previous outputs are preserved.
Interesting Challenges & Approaches
Through the process of designing ADAMS, we arrived at several guiding principles. First, modules should be robust and well-tested before integrating them into any agentic system, as what initially appeared to be agent limitations often turned out to be gaps in module reliability. Second, key entry points should be encapsulated and smaller, focused agents should be built first rather than monolithic ones. Third, consistent log messages across the entire pipeline can help the agents more effectively debug themselves. Finally, development should start with verbose prompts and then iteratively trim them down to core principles, and the essential insights from the detailed versions will persist.
One of our key technical challenges was determining the appropriate degree of autonomy for the agents. If the pipeline is too rigid and standardized, with agents limited to tool calling through pre-defined entry points, any user request that falls outside the established scope will be rejected. Conversely, if agents are granted too much freedom, uncertainty increases and it becomes harder to ensure that proposed execution plans align with both the user's intent and the available tool capabilities. We believe that striking the right balance between structured tool calling and flexible execution is the key to constructing a truly agentic workflow.
Future Work
We will continue to make further improvements on ADAMS in 2026. We plan to expand the number of tools being used and have an interesting R&D roadmap for making multi-agent systems be useful for discoveries in chemistry. We hope this kind of work pushes the field forward in realizing what's possible in this new age for scientific research.
Citation
@software{adams2025,
title = {ADAMS: Agent-Driven Autonomous Molecular Simulations},
author = {Rhizome Research},
year = {2025},
url = {https://github.com/Applied-Scientific/ADAMS},
license = {Apache-2.0}
}You can contact us at x@rhizome-research.com, fill out our form here or find us on LinkedIn for partnership opportunities.